Post by: JulesMhz on May 17, 2023, 10:54:11 AM
I have a question regarding molecule similarity computation. I'm more from computation than chemistry, so it is a fairly new topic for me, and I'm actually working with a (quantum computing) algorithm for molecule similarity computation.
So here is my question, given these molecules:
- niacin
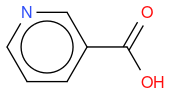 , hereafter "reference molecule"
, hereafter "reference molecule"- 4-CARBOXYPIPERIDINE
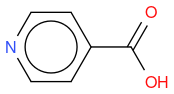 , hereafter "molecule 1"
, hereafter "molecule 1"- nicotinamide
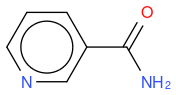 , hereafter "molecule 2"
, hereafter "molecule 2"- P modified nicotinamide
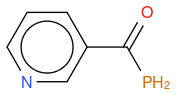 , hereafter "molecule 3"
, hereafter "molecule 3"If I compute the Tanimoto similarity between reference and molecule 1, I have 0.419.
If I compute the Tanimoto similarity between reference and molecule 2, I have 0.633.
What I observe is that Tanimoto similarity considers that molecule 2 is more similar to reference molecule than molecule 1, but if we look at molecule illustrations, we notice that molecule 1 differs from reference by one N atom moved by one position, whereas molecule 2 differs from reference by one molecule which is not the same.
So, in an algorithmic point of view, it makes sense that molecule 1 has two molecule differences (one N replaced by C, and one C replaced by N) whilst molecule 2 has only one molecule difference (OH replaced by NH2) so the similarity is lower for molecule 1.
But, in a chemical point of view, does this also make sense ? I mean, why just moving one N atom is less similar than changing one atom by an other ? In other word, is the chemical function of molecule 2 more similar than molecule 1 to reference molecule ?
An other observation, if I compute the Tanimoto similarity between reference and molecule 3, I have 0.633 (like for molecule 2), so Tanimoto distance does not take in account the fact that one atom differs between molecule 2 and 3, whilst my "non-chemical-specialist" mind would guess than one is more similare than the other as they are not equivalent ?
Finally, is there a "chemical" process (by chemical, I mean not algorithmic) to compare molecules in order to have "chemical function" similarity I can refer to ?
Thank you for your help, I hope my questions are well formulated.
Post by: Borek on May 17, 2023, 02:23:17 PM
What is more similar to a square - a triangle, or a pentagram?
Sure, you can choose some set of rules to calculate "similarity" index, but it will be always arbitrary and as such can work for some applications and not work for others. This is a can of worms if you want to pretend there is any strict science behind.
Post by: Corribus on May 22, 2023, 02:23:16 PM
But, in a chemical point of view, does this also make sense ? I mean, why just moving one N atom is less similar than changing one atom by an other ? In other word, is the chemical function of molecule 2 more similar than molecule 1 to reference molecule ?It would also be important to address the question: similar with respect to what? 1 and 2 may be, for example, the most similar in terms of boiling point, but the least similar in terms of reactivity with an acid. (Just a hypothetical, I didn't look anything up.) You need to define your property of interest; similarity between chemical structures means nothing in any general sense.
Post by: clarkstill on May 24, 2023, 09:23:17 AM